Open database encapsulates worldwide knowledge of human metabolism
An international research consortium, spearheaded by the Luxembourg Centre for Systems Biomedicine (LCSB) of the University of Luxembourg, has developed a database that is unique in the world: the Virtual Metabolic Human Database (VMH). VMH is a collection of knowledge that researchers from all around the world have generated about human metabolism in the last 60 years.
Responsible for the project is Prof. Ines Thiele, head of the LCSB Molecular Systems Physiology group. Thiele and her team together with project partners from France, the Netherlands and India have systematically collected and aggregated data on human metabolism, diseases, microbial gut flora, and nutrition. The researchers have also combined this aggregated knowledge with comprehensive computer models of human metabolic processes. The VMH database is an open source project. Open source means that the results of publicly funded research have been made available to the general public. “Thus, every detail of research into metabolism is just a mouse click away for any researcher anywhere,” says Ines Thiele. The scientific journal Nucleic Acids Research published the results on VMH on October 29 as a special interest “breakthrough” article.
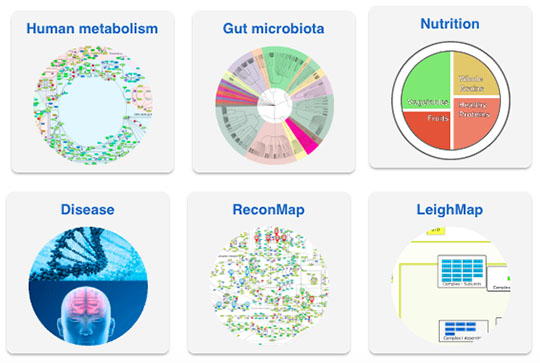
Few human diseases have a single trigger as their cause – like a single mutated gene, for example. Instead, most of the time, there are countless internal and external influences at play. That is exactly how it is for complex diseases, such as Parkinson’s, Alzheimer’s, and cancer. While researchers have discovered a multitude of details on disease patterns, “until now this knowledge has been scattered across different papers and databases,” says Ines Thiele. “Even for specialists, it is difficult to see the full picture.” Accordingly, unless they just so happened to be published at the same time or in the same journal, retrieving existing correlations between research results is time-consuming and cumbersome. That is set to change with the VMH.
A unique database on human metabolism
With the public release of the VMH, the research consortium led by Ines Thiele has brought a temporary end to a truly herculean task. For five years, the researchers collected and aggregated data available from publications and databases on the topic of metabolism. These data had to be curated, meaning they had to be prepared to be compatible for use in the VMH. The research team has also developed a so-called Nutrition Designer for data obtained from nutrition research. This Designer can be used to run simulations of the metabolic processes with a focus on diet. Thiele remarks, “it’s something entirely new and pretty cool.” This effort was all made possible by the funding obtained for the project through Ines Thiele’s ATTRACT fellowship from the Luxembourg National Research Fund (FNR), her ERC Starting Grant, and the NCER-PD project also funded by the FNR.
Young students involved in the project
What is very special about the VMH is that secondary and tertiary-level students were included in the creation of the database, as project partner Prof. Ronan Fleming of the University of Leiden in the Netherlands and former head of the LCSB Group “Systems Biochemistry” explains: “Last year, we hosted a summer course for school and tertiary students together with the Scienteens Lab of the University of Luxembourg. In this course, the students worked together on preparing VMH data and visualising them in a google-like map of human metabolism.” Even though most of the students had little or no prior scientific experience, they made a highly valuable contribution to the project, as Fleming relates: “Everyone who worked on this project is therefore also a co-author of the paper.”

L’équipe du projet MitoCarte – The MitoCarte team
An efficient tool for biomedical research
The VMH will provide a considerable boost to biomedical work, Ines Thiele is convinced: “Our database reveals hitherto hidden connections between processes of human and gut microbial metabolism.” Among other things, this is important for the development of active pharmaceutical ingredients (APIs). For instance, it is now possible to ascertain whether a drug aimed at fighting a certain disease might also upset gut bacteria that are physiologically important to our metabolism. Thiele continues, “With one click, VMH tells me whether the human metabolic pathway or the protein targeted by the API is also present in any bacteria.” Then, by cleverly modifying that API, it may be possible to avoid side effects, for example.
For the researchers, the release of the VMH does not mark the end of their work, as Thiele explains: “We have to intensify our efforts even further, so that the VMH is remains up to date with new research insights and becomes a truly effective work tool for biomedical research.” The next stage is acquiring funding to hire more scientists and programmers. “Now that VMH is online, we will be receiving all kinds of feedback and pointers to more data from research colleagues,” Thiele says. “The integration of this information into the VMH will involve substantial efforts.” Thiele envisages to expand the VMH to become a predictive simulation tool. “Then it will be possible to analyse even more aspects of human metabolism and our environment – and eventually to make personalised predictions, or in other words predictions for a specific patient, if the corresponding data exist.”
—
Financing:
This study received funding from the Luxembourg National Research Fund (FNR) through the ATTRACT program (FNR/A12/01), the CORE program (C16/BM/11332722), the OPEN grant (FNR/O16/11402054), the National Centre of Excellence in Research (NCER) on Parkinson’s disease, the European Union’s Horizon 2020 research and innovation program under grant agreement No. 668738 and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 757922). This study received generous support from the Art2Cure ASBL for the generation of ReconMaps.
Bibliography:
Alberto Noronha, Jennifer Modamio, Yohan Jarosz, Elisabeth Guerard, Nicolas Sompairac, German Preciat, Anna Dröfn Daníelsdóttir, Max Krecke, Diane Merten, Hulda S Haraldsdóttir, Almut Heinken, Laurent Heirendt, Stefanía Magnúsdóttir, Dmitry A Ravcheev, Swagatika Sahoo, Piotr Gawron, Lucia Friscioni, Beatriz Garcia, Mabel Prendergast, Alberto Puente, Mariana Rodrigues, Akansha Roy, Mouss Rouquaya, Luca Wiltgen, Alise Žagare, Elisabeth John, Maren Krueger, Inna Kuperstein, Andrei Zinovyev, Reinhard Schneider, Ronan M T Fleming, Ines Thiele; The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease, Nucleic Acids Research, gky992, https://doi.org/10.1093/nar/gky992
