The University’s High-Performance Computing (HPC) facility has significantly contributed to the fight against the COVID-19 pandemic since mid-March 2020.
The “supercomputer” and its team, led by Prof. Pascal Bouvry and Dr. Sébastien Varrette, has supported University researchers and external partners in more than seven projects with its computational resources.
HPC delivers high performance in order to solve large problems faster. Tasks which would typically require several years to be computed on a typical desktop computer may only require a couple of hours, days or weeks over an HPC system. And for research conducted to fight the COVID-19 pandemic, accelerating time-to-solution is a critical criterion to efficiently fight the spread of the pandemic.
The University supercomputer’s vast computing power and storage capacities were used for enabling and accelerating COVID-19 research in the areas of biomedical & life sciences, ICT and material sciences. Among others, it facilitates machine learning-based lung bioavailability estimations; business ecosystem modelling and simulation techniques to inform economic policy-makers in Luxembourg and abroad; while allowing for computing future predictions of the visibility of the coronavirus on surfaces.
The University’s HPC services support four University-led projects funded by the FNR COVID-19 Fast Track Call, one project from the Research Luxembourg COVID-19 Task Force and one collaboration between the University’s Luxembourg Centre for Systems Biomedicine, TU Munich and the Flatiron Institute.
The HPC facility is one element of the extensive digital research infrastructure and expertise developed by the University over the last years. It also supports the University’s ambitious digital strategy and in particular the creation of a Facility for Data and HPC Sciences. This facility aims to provide a world-class user-driven digital infrastructure and services for fostering the development of collaborative activities related to frontier research and teaching in the fields of Computational and Data Sciences, including High Performance Computing, Data Analytics, Big Data Applications, Artificial Intelligence and Machine Learning.
More than 1030 jobs were scheduled on the dedicated reservations set by the HPC team (the longest job running for 58 days). An overview of the associated load usage in the most critical period of the pandemic is depicted in Figure 1. The University’s HPC remains determined to provide resources and guidance to current and future COVID-19 related projects.
Graphique 1 : Aperçu des ressources de calcul utilisées par les projets COVID-19 de mi-mars à fin avril. 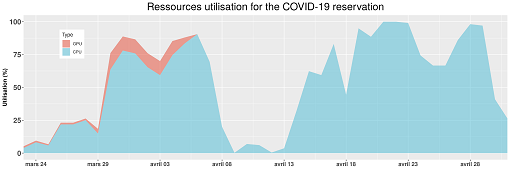
The high utilisation rate of the resources during this critical period shows the strong involvement and collaboration of all University partners to fight this pandemic. Below is a list of the main COVID-19 related projects which relied on the University’s HPC computing resources:
Combined In Silico Molecular Docking And In Vitro Experimental Assessment Of Drug Repurposing Candidates For Covid-19 (CovScreen)
Currently, no vaccine or sufficiently validated pharmacological treatment is available for COVID-19. Drug-based strategies to reduce the viral load in patients with severe forms of COVID-19 include the repurposing of existing small molecule compounds that inhibit the activity of key viral proteins or human proteins involved in mediating viral entry or release from the host cell. The project proposes a combined computational and experimental approach to rank an alternative candidate known as drugs, antivirals, and natural compounds, which are commercially available, inexpensive, and safe in humans. The project screens and filters in silico x M~10k compounds using molecular docking and machine learning based lung bioavailability estimations and conducts molecular dynamics simulations for refined binding affinity estimation of the 100 top-ranked compounds. The project aims to provide a fast experimental validation of drug repurposing candidates for COVID-19 from a computational pre-selection of antivirals, drugs, and natural compounds that are inexpensive, have known safety properties, and high predicted bioavailability in the lung. The project is led by Enrico Glaab.
Figure 2: A) Modeled SARS-CoV-2 Spike Glycoprotein overlaid with the SARS-CoV (PDB: 2GHV) unique amino acids are shown. Variable amino acid residue side chains are shown: Green: SARS-CoV Red: SARS-CoV-2. B) Minimised final structure of modeled SARS-CoV-2 spike glycoprotein. Source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7152904/figure/fig1/ 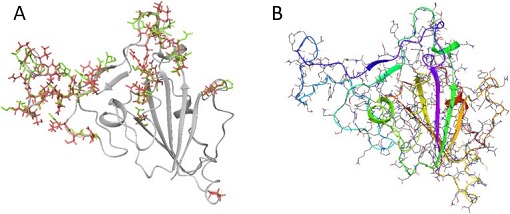
Machine Learning To The Rescue: From Health Recovery To Economic Revival (REBORN)
REBORN is a data science project that focuses on ensuring sustainable economic recovery in the face of COVID-19. The project team applies advanced Machine Learning, business ecosystem modelling (i.e., expert knowledge) and simulation techniques to yield recommendations of economic actions given different scenarios in which the lockdown is relaxed, partially or totally lifted. By interacting with other teams of the Luxembourg Task Force, this project targets high impact for the various sectors of the Luxembourg economy, by providing appropriate data-driven recommendations for political decision-makers. REBORN aims to answer important questions such as: What are the industrial sectors to help in priority? What are the sectors that should be restarted first? What are the possible changes in consumer habits and the impact of neighboring country decisions on commuters? etc. Ultimately, REBORN contributes towards reflections on initiatives to limit the spread of the future economic crisis due to COVID-19 as well as avoid worsening future waves of coronaviruses. The project is led by Jacques Klein.
Figure 3: Tools to strengthen resilience for COVID-19. Source: https://council.science/current/blog/setting-up-a-data-ecosystem-to-defeat-covid-19/ 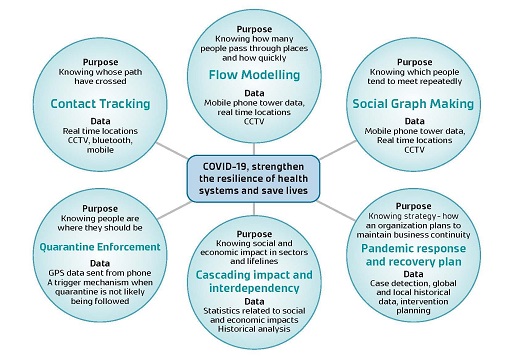
Privacy Preserving Monitoring of Social Distancing In Public Environments Machine Learning, Computer Vision, Social Distancing, GDPR By Design (PEOPLE)
The project aims to provide a platform to run a comprehensive analysis on the Social Distancing measures decided by the government in the context of the COVID-19 pandemic. It aims to analyse anonymised video data in the city of Luxembourg. The first step is to anonymise the video feed using well-known Artificial Intelligence (AI) models (face blurring). The next step uses other AI models to identify pedestrians and groups of individuals, calculate their relative distances and overall density. Those metrics can then be evaluated over time for different locations and provide valuable insights on the greater or lesser risks of infection spreading based on behaviour. The rules can be used to inform where the police need to focus their efforts on enforcing laws or informing and influencing the public’s actions (or both). The project is led by Raphael Frank.
Virus-surface Interactions In Dynamic Environments (V-SIDE)
The scientific community is trying to establish how long COVID-19 can survive on a given surface (e.g., paper, plastics, glass, and metals). Their properties (smooth v/s rough) play a crucial role during the infection spreading phase. Using the Fourier-transform infrared spectroscopy (FTIR) helps to predict the surface-specific visibility of the coronavirus in the steady-state and dynamic state (e.g., changes in temperature and humidity). Later the obtained model will be mapped into the biophysical model system. Machine learning techniques are used for an optimistic and future prediction of the visibility of the virus on the surfaces. This is the part where the University’s HPC software and hardware facilities will be used. The project is led by Anupam Sengupta.

Figure 4: How long does the COVID-19 virus survive on surfaces?Source: New England Journal of Medicine
Sars-CoV-2 protein structure prediction
Bioinformaticians from Rostlab at TU Munich, the LCSB and the Flatiron institute joined forces to participate in a worldwide effort to predict 3D structures of Sars-CoV-2 proteins. The project was organised by the team behind CASP (Critical Assessment of protein Structure Prediction), who selected ten viral proteins for which no experimental structure is available, nor can homology-based modeling be used to infer structure. The goal is to obtain consensus structures, which would give insight into the molecular mechanisms of the virus, as well as aid vaccine development and the evaluation of possible drug targets.
To this end, the team from TUM and LCSB, helped by collaborators at the Flatiron Institute, trained multiple Deep Learning systems, leveraging evolutionary information from multiple sequence alignments, to predict pairwise distances of amino acids in proteins. The hardest targets of previous CASP competitions were used to assess the reliability of predictions. Computer generated distance maps were then used as constraints to simulate protein folding and obtain 3D structures. Three nodes of the Iris HPC cluster with four Tesla GPUs each (in addition to three similar nodes at the Flatiron Institute and one node from the UCC at TUM) were used to prepare datasets, as well as to train DL models, enabling high quality submissions within the initiative’s short-term.
Figure 5: (left) Atomic-level structure of the spike protein of the virus that causes COVID-19 (Source: McLellan Lab, University of Texas at Austin); (right) Visualisations of predicted protein structures from this project. 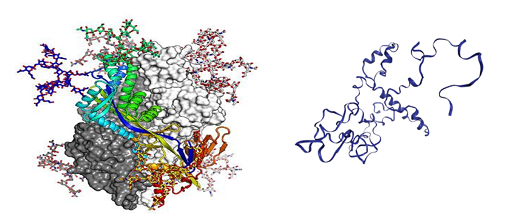
COVID-19 Task Force
In response to the global COVID-19 pandemic, the COVID-19 Task Force was set up by Research Luxembourg at the early stage of the confinement. As part of Work Package 6 (statistical pandemic projections), researchers from LCSB have developed a computational agent-based model that is essential in understanding the progression of the Covid-19 epidemic in Luxembourg. The statistical projections are required to avoid saturation of the healthcare system and can be used to aid in political decision-making by simulating the epidemic development and its associated outcomes under various future scenarios and strategies.
In order to make informed decisions, hundreds of scenarios with several randomised replicates need to be considered. Parallelisation of the underlying computer code has been key to speed up the simulations and reduce the computing time to an absolute minimum. Thanks to the HPC platform of the University of Luxembourg and its team, it is now possible to launch hundreds of scenarios simultaneously, leading to results being made available to decision-makers within minutes.
Medical image analysis of X-ray and CT of COVID19
During the past weeks, the HPC infrastructure and in particular the GPUs were employed for the AICovIX project. This project focuses on the development of automated solutions for the analysis of medical image analysis of X-ray and CT of COVID19 pneumonia patients using computer vision and deep learning techniques. This work is conducted by researchers of the INS group.
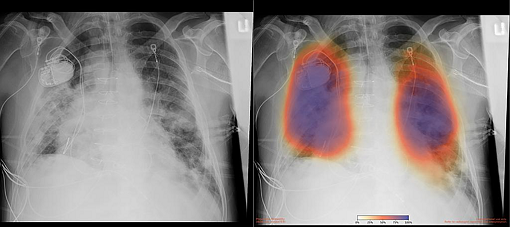
Figure 6: Chest X-rays from a patient with COVID-19 pneumonia, original x-ray (left) and AI-for-pneumonia result (right) (Photo courtesy of UC San Diego Health).
