A glimpse into our research
Our group helps with handling, storing and categorising data, and providing automatic workflows that apply prediction, filtering, and summarisation steps in order to focus on the relevant aspects of the data. Computer scientists and biologists covering a broad spectrum of disciplines work together in order to develop and deploy time- and cost-efficient analysis pipelines.
Supporting research and operations at the LCSB
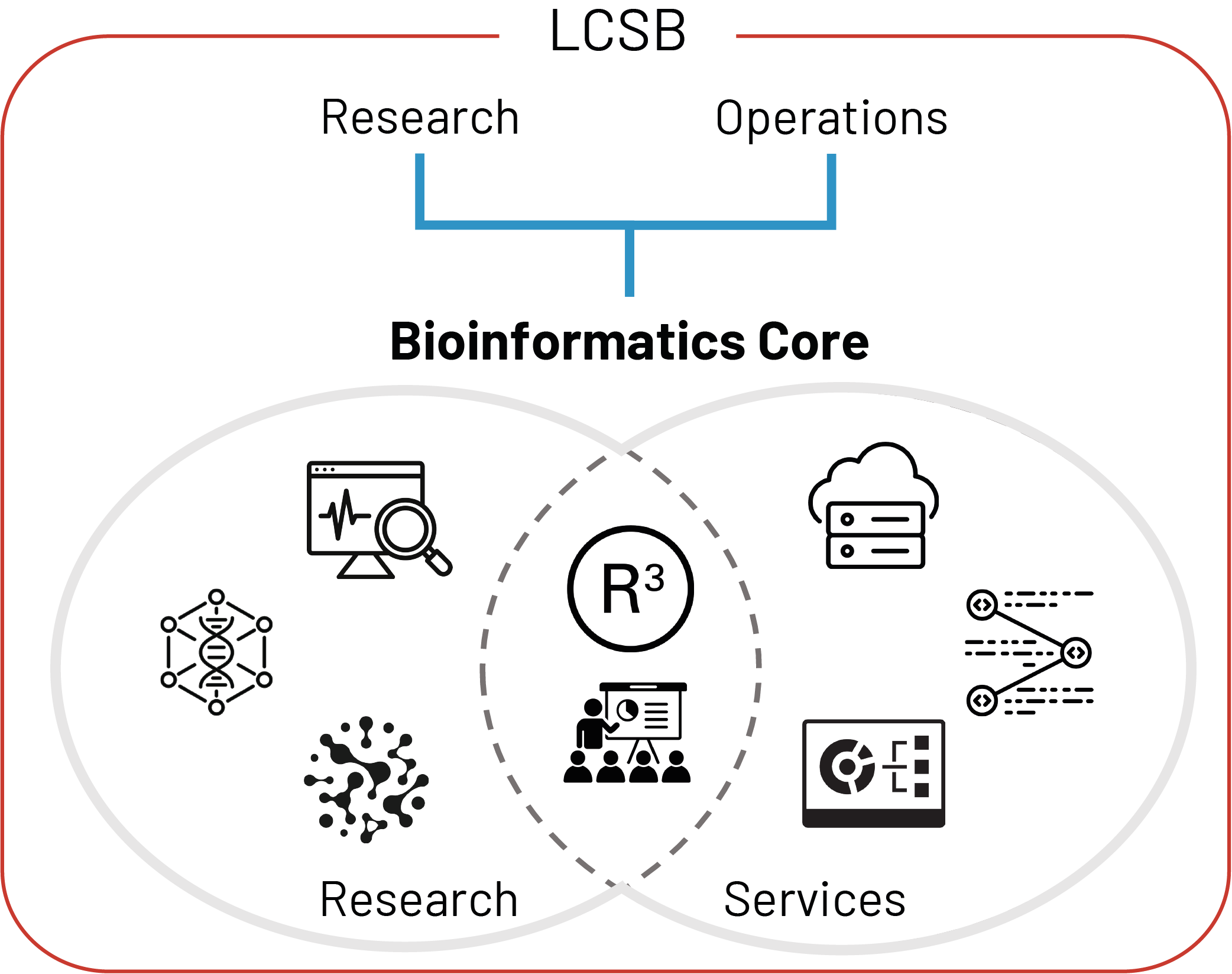
Our research involves
– genomics
– translational medicine
– visual analytics
We offer services in
– data management and stewardship
– custom tool development
– research IT infrastructure
Reproducible research and training span research and services

